Analysis
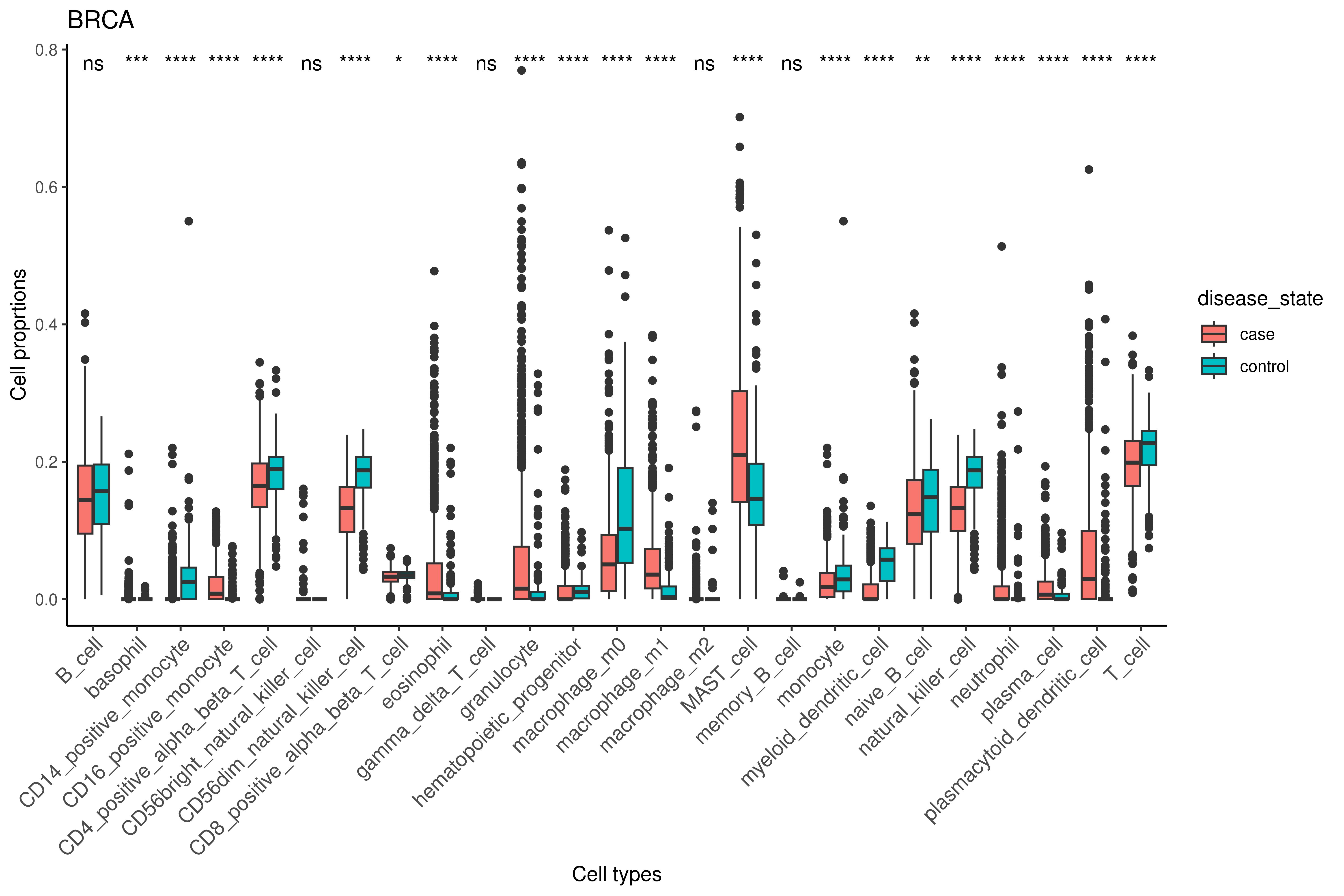
Cell Types Analyses
Cell-mixture Deconvolution
DescriptionCell-mixture Deconvolution
To estimate cellular proportions, we applied a computational deconvolution method using gene expression profiles from bulk tissue samples.
The core of this approach is the use of a pre-defined "basis matrix", which contains gene expression profiles of specific cell types and their corresponding marker genes. These marker genes, which are either uniquely or predominantly expressed by particular cell types, serve to identify and quantify the relative abundance of each cell type in the mixed sample.
We used the ImmunoStates basis matrix from the MetaIntegrator R package, which includes profiles for 20 immune cell types, to estimate the proportions of these cell types in our samples.
Mathematically, the deconvolution process involves solving a linear regression model, where the observed bulk expression values are modeled as a weighted sum of the reference expression profiles in the basis matrix. The weights correspond to the estimated proportions of each cell type, providing a cellular composition profile for each sample.
Example: